Rhodamine Labeling Strategies Designed for Your Specific Biomolecules
Rhodamine labeling, as a key fluorescence labeling technique for biomolecules, has become an indispensable tool in cell imaging, protein localization, nucleic acid analysis, and molecular interaction studies due to its high brightness, excellent photostability, and broad applicability. Through customized Rhodamine labeling strategies tailored for proteins, antibodies, nucleic acids, peptides, and small-molecule probes, researchers can achieve high-sensitivity detection, precise visualization, and multichannel colocalization analysis, enabling in-depth investigation of dynamic processes and molecular mechanisms within complex biological systems.
Understanding Rhodamine Labeling in Biomolecular Research
In modern molecular biology and biochemistry research, fluorescence labeling technologies have become essential tools, and Rhodamine fluorophores occupy a prominent position in biomolecular labeling due to their superior optical properties and stability. Through Rhodamine labeling, researchers can sensitively detect and track biomolecules such as proteins, nucleic acids, and antibodies, providing powerful support for cell imaging, protein localization analysis, and studies of molecular interactions.
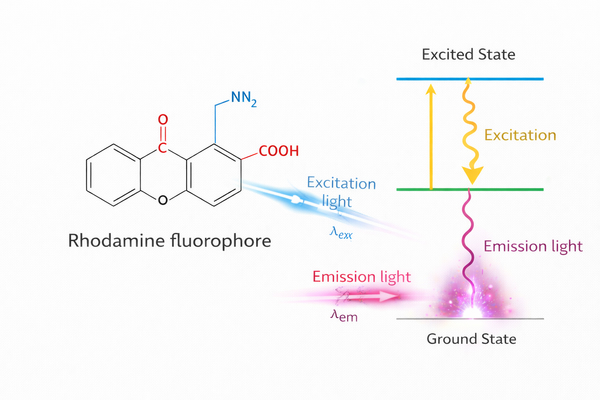 Fig. 1. Fluorescence mechanism and spectral properties of rhodamine fluorophores used in biomolecular labeling (BOC Sciences Authorized).
Fig. 1. Fluorescence mechanism and spectral properties of rhodamine fluorophores used in biomolecular labeling (BOC Sciences Authorized).
Fundamental Properties of Rhodamine Fluorophores for Bio-labeling
From a chemical structure perspective, the Rhodamine family is characterized by a planar conjugated system composed of a xanthene core and three aromatic rings.
- High molar extinction coefficient and quantum yield: The rigid ring structure effectively suppresses non-radiative decay pathways (vibrational relaxation) in the excited state, resulting in a molar extinction coefficient (ε) typically exceeding 80,000 M⁻¹ cm⁻¹. This allows Rhodamine dyes to capture sufficient photons even in extremely low-concentration biological samples.
- Biological significance of pH stability: Unlike FITC (Fluorescein Isothiocyanate), whose fluorescence is highly sensitive to environmental pH and is severely quenched at pH < 6, Rhodamine fluorescence remains highly stable across the physiological pH range of 4.0 to 9.0. This property makes Rhodamine an ideal choice for studying biological events in lysosomes (acidic environments) or mitochondria (alkaline environments).
- Flexibility of amino substitution: Substitution at the 3- and 6-amino positions of the xanthene ring (e.g., alkylation or ring closure) allows precise tuning of excitation and emission wavelengths, giving rise to red-shifted derivatives such as Sulforhodamine 101.
Why Rhodamine Dyes are Preferred for High-Sensitivity Detection?
In high-sensitivity detection applications, the signal-to-noise (S/N) ratio is the ultimate criterion for evaluating labeling strategies.
- Photostability: Rhodamine dyes exhibit intrinsic resistance to photo-oxidation. Under continuous high-intensity laser scanning in confocal microscopy, Rhodamine fluorescence shows a significantly longer half-life than fluorescein. This property is critical for super-resolution microscopy techniques such as STED, which require extremely high depletion laser power to compress the point spread function (PSF).
- Red-shifted spectra to avoid background interference: Endogenous biomolecules in biological samples (e.g., NADH, flavoproteins) exhibit strong autofluorescence in the blue-green region (400–500 nm). Rhodamine emission is primarily distributed between 550 and 650 nm, effectively avoiding this noisy spectral region and ensuring high-contrast imaging.
- Tunable Stokes shift: Structural modifications (e.g., Texas Red) can increase the Stokes shift, reducing spectral overlap between excitation and emission light and further minimizing detection interference.
Looking for Rhodamine Dyes?
Whether you require specialized Rhodamine derivatives, water-soluble variants, or photostable forms, our experts offer tailored synthesis solutions to meet your research and experimental needs.
Biomolecules Suitable for Custom Rhodamine Labeling
Rhodamine dyes exhibit broad applicability and enable efficient, stable fluorescence labeling of various biomolecules. By tailoring Rhodamine labeling strategies to the structural and functional characteristics of different biomolecules, specificity, signal intensity, and biological activity can be preserved, providing reliable support for diverse research applications.
Rhodamine Labeling of Proteins and Enzymes
Proteins and enzymes are among the most commonly labeled targets in life science research. Rhodamine dyes can be covalently conjugated to amino or cysteine residues on proteins, enabling stable labeling. By optimizing labeling conditions, high signal intensity can be achieved while preserving enzymatic activity and protein conformation. Rhodamine-labeled proteins and enzymes are widely used in protein quantification, enzyme kinetics studies, protein–protein interaction network analysis, and drug screening assays.
- Non-specific surface labeling: Modification of ε-amino groups on lysine residues using NHS esters or isothiocyanates (TRITC), suitable for tracing experiments requiring high fluorescence intensity.
- Site-specific modification: To avoid interference with active sites, cysteine residues located in non-functional regions are often selected and conjugated using maleimide chemistry.
- Preservation of enzymatic activity: When labeling enzymes, the dye-to-protein molar ratio (F/P ratio) must be carefully controlled. Excessive dye loading may cause protein misfolding or aggregation due to hydrophobic Rhodamine stacking.
Custom Rhodamine-Labeled Antibodies
Antibody labeling is a core technique in immunological research and protein detection. Rhodamine-labeled antibodies enable high-sensitivity detection and visualization of protein localization. By selecting appropriate conjugation chemistries and controlling the degree of labeling, stable and quantifiable fluorescence signals can be obtained while maintaining antibody specificity. Labeled antibodies are widely used in immunofluorescence imaging, colocalization analysis, multiplex fluorescence detection, and flow cytometry.
- Fc-directed labeling: Oxidation of antibody glycans generates aldehyde groups that can be conjugated with Rhodamine hydrazides, effectively protecting antigen-binding (Fab) regions.
- Secondary antibody conjugation optimization: In multicolor immunofluorescence, Rhodamine dyes (e.g., Texas Red) are commonly paired with green fluorophores. Precise chemical conjugation ensures high signal-to-noise ratios in complex cellular matrices.
- Non-specific lysine labeling: Suitable for routine fluorescence labeling, providing uniform and high-intensity signals.
- Cysteine site-specific labeling: Applied to monoclonal antibodies or Fab fragments to ensure labeling sites are distant from antigen-binding regions, preserving antibody activity.
Rhodamine Labeling of Peptides
Peptides play critical roles in signal transduction, receptor–ligand studies, and drug screening. Rhodamine dyes can be site-specifically conjugated to the N-terminus, C-terminus, or side-chain functional groups of peptides, ensuring labeling stability and positional control. Labeled peptides are used for intracellular tracking, receptor binding studies, and dynamic behavior analysis, providing reliable tools for drug discovery and mechanistic research.
- Terminal labeling: Labeling at the N- or C-terminus preserves peptide function while providing stable and uniform fluorescence signals for cell tracking or binding assays.
- Side-chain labeling: Selective conjugation via lysine or cysteine side chains enables site-specific labeling for mechanistic studies or target-binding experiments.
- Activity preservation: Controlling dye molar ratios and reaction time prevents conformational changes or aggregation, ensuring reproducibility and reliability.
Rhodamine Labeling of Nucleic Acids and Oligonucleotides
Nucleic acids and oligonucleotides are central molecules in gene expression analysis, probe development, and molecular detection. Rhodamine labeling can be covalently introduced at the 5′ end, 3′ end, or internal bases to form stable fluorescent probes. Labeled nucleic acids are used in real-time quantitative PCR, fluorescence in situ hybridization (FISH), RNA tracking, and molecular recognition studies, enabling high-sensitivity detection and quantitative analysis.
- Terminal labeling: 5′ or 3′ end conjugation is suitable for qPCR, FISH, and RNA tracking, providing clear signals and high sensitivity.
- Internal labeling: Conjugation at specific bases or non-binding regions enables conformational studies and analysis of molecular interactions while minimizing functional interference.
- Stability optimization: Label design should consider secondary structures and buffer conditions to ensure stable and specific fluorescence under experimental conditions.
Rhodamine Labeling of Aptamers
Aptamers are highly specific molecular recognition tools. Rhodamine labeling enables visualization without compromising binding activity. Labeled aptamers are used in target capture, cell surface receptor detection, and high-throughput screening, facilitating studies of molecular recognition mechanisms and optimization of probe design.
- Terminal conjugation: Labeling at the N- or C-terminus avoids interference with binding regions and improves signal reproducibility.
- Internal modification: Labeling at non-binding regions enhances fluorescence without affecting conformation, suitable for colocalization analysis or high-throughput screening.
- Retention of binding activity: Adjusting dye molar ratios ensures labeling does not alter aptamer folding or target affinity.
Rhodamine Labeling of Receptors and Ligands
Receptors and ligands play key roles in cell signaling and drug research. Rhodamine labeling enables tracking of receptor–ligand interactions, analysis of binding kinetics, and investigation of signaling pathways. Fluorescence-based monitoring allows precise analysis of receptor distribution, activity changes, and ligand mechanisms of action.
- Lipophilicity balance: Due to the inherent lipophilicity of Rhodamine, hydrophilic linkers are often introduced when labeling hydrophobic ligands to maintain solubility in physiological buffers.
- Pharmacological activity validation: Labeled ligands should be validated through saturation binding assays to confirm that dissociation constants (Kd) remain consistent with unlabeled molecules.
- Receptor labeling: Dye conjugation via reactive functional groups enables subcellular localization and dynamic tracking for signaling pathway and target analysis.
- Ligand labeling: Labeling sites are selected to avoid affecting binding capability, supporting binding assays, competition studies, and kinetic measurements.
Rhodamine Labeling of Small-Molecule Probes
Small-molecule probes are important tools in cell imaging, signaling pathway analysis, and pharmacokinetic studies. Rhodamine conjugation enables high-brightness fluorescence output in physiological environments, facilitating tracking of intracellular distribution and molecular behavior and providing visual support for mechanistic studies.
- Photo-click chemistry applications: By introducing photoactivatable groups (e.g., azides), Rhodamine can be used for capture labeling to study intracellular target distribution of drug molecules.
- Chemical site selection: Conjugation via hydroxyl, amino, or other reactive functional groups ensures stable and specific labeling.
- Functional preservation: Labeling does not interfere with probe targeting capability or activity.
- Fluorescence optimization: Adjusting dye loading and reaction conditions achieves optimal signal intensity, stability, and experimental reproducibility.
Rhodamine Labeling of Glycans and Carbohydrates
Glycans and polysaccharides are critical in cell surface recognition, glycoprotein analysis, and signal transduction studies. Rhodamine labeling enables stable conjugation via hydroxyl, amino, or other functional groups, allowing visualization and quantitative detection. Labeled glycans are used in cell surface glycan analysis, glycoprotein recognition studies, and investigation of glycosylation-related signaling pathways.
- Metabolic labeling: Cells uptake azide-containing sugar precursors, followed by intracellular click chemistry to conjugate alkyne-modified Rhodamine to cell surface glycans.
- Reductive amination: For isolated glycans, labeling via aldehyde groups at the reducing end is a classical characterization method.
- Functional group selection: Conjugation via hydroxyl, amino, or other reactive groups enables stable labeling.
- Stable labeling: Ensures long-term fluorescence stability in aqueous or physiological conditions with minimal photobleaching.
Custom Rhodamine Conjugation Strategies and Chemistry
In biomolecular labeling, selecting appropriate Rhodamine conjugation strategies is critical for ensuring labeling efficiency, fluorescence signal stability, and preservation of biological activity. Different biomolecules possess distinct structures and chemical properties, requiring tailored labeling approaches. Well-designed conjugation strategies not only enable precise control of labeling sites and labeling degree but also minimize nonspecific binding and functional loss, thereby supporting complex downstream analyses and high-precision experimental applications.
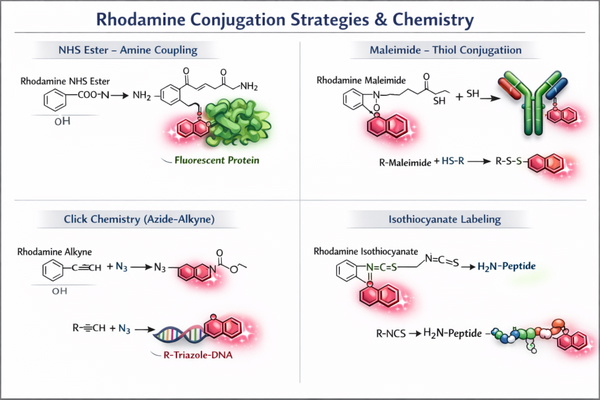 Fig. 2. Rhodamine conjugation strategies via NHS ester, maleimide, click chemistry, and isothiocyanate (BOC Sciences Authorized).
Fig. 2. Rhodamine conjugation strategies via NHS ester, maleimide, click chemistry, and isothiocyanate (BOC Sciences Authorized).
Amine-Reactive Rhodamine Labeling for Proteins and Antibodies
Amine-specific labeling is one of the most widely used Rhodamine conjugation strategies. It typically involves NHS ester or isothiocyanate derivatives reacting covalently with lysine residues on proteins or antibodies. This approach features mild reaction conditions, high efficiency, and stable labeling. During the labeling process, pH and dye molar ratios must be optimized to control the degree of labeling and to avoid excessive modification that may cause protein conformational changes or reduced antibody activity. Amine-reactive Rhodamine labeling is widely applied in fluorescence immunoassays, protein quantification, enzyme activity studies, and multichannel imaging experiments.
- NHS ester strategy: This is currently the most stable conjugation chemistry. Under mildly alkaline conditions (pH 8.2–8.5), Rhodamine–NHS esters undergo nucleophilic substitution with primary amines to form highly stable amide bonds.
- TRITC (isothiocyanate) strategy: Rhodamine isothiocyanates conjugate via thiourea bond formation. Although their reactivity is slightly lower than that of NHS esters, the chemical properties of thiourea linkages can provide distinct solubility characteristics in specific proteomics applications.
- Key factors in reaction control: Since multiple lysine residues are typically present on protein surfaces, controlling dye-to-protein stoichiometry is essential to prevent over-labeling and protein precipitation caused by increased hydrophobicity.
Thiol-Specific Rhodamine Conjugation for Site-Selective Labeling
For applications requiring highly site-specific labeling, cysteine thiol groups are ideal targets. Thiol-specific conjugation provides superior control over labeling sites and enables precise site-selective modification. Common strategies include maleimide chemistry or reduced disulfide-based conjugation. This approach is particularly suitable for function-sensitive proteins and antibodies, ensuring that critical active sites remain unaffected. Cysteine-specific conjugation yields more homogeneous labeled products, improves fluorescence signal consistency, and reduces the risk of nonspecific binding.
- Maleimide chemistry: Under near-neutral conditions (pH 6.5–7.5), Rhodamine–maleimide reacts with thiols via a Michael addition. This reaction exhibits extremely high chemical selectivity, as amine deprotonation and reactivity are minimal within this pH range.
- Advantages of site specificity: Introducing a single cysteine residue at a defined position through genetic engineering enables precise “one dye–one site” Rhodamine labeling. This is critical for single-molecule fluorescence studies, as it ensures that each detected photon signal corresponds to a defined molecular conformation.
Click Chemistry and Advanced Coupling Approaches for Precision Labeling
Click chemistry enables covalent conjugation of Rhodamine to biomolecules through alkyne–azide reactions. This method supports precise labeling in complex systems while maintaining high yields across diverse functional group environments. Click chemistry is particularly well suited for multiplex labeling experiments and advanced probe design, offering rapid, controllable, and highly biocompatible conjugation. Applications include nucleic acid labeling, multichannel protein imaging, and dynamic molecular tracking studies.
- CuAAC (copper-catalyzed azide–alkyne cycloaddition): Azide-functionalized biomolecules react with terminal alkyne-modified Rhodamine. This reaction is irreversible, high-yielding, and extremely fast in aqueous environments.
- SPAAC (strain-promoted azide–alkyne cycloaddition): Using strained cyclooctynes (e.g., DBCO), labeling proceeds without toxic copper catalysts. This represents a major advance for in situ Rhodamine labeling in live cells, significantly reducing cytotoxicity.
- Tetrazine–trans-cyclooctene (TCO) coupling: One of the fastest bioorthogonal reactions known, making it particularly suitable for dynamic Rhodamine labeling at very low concentrations or with short-lived isotopes.
Controlling Labeling Degree and Preserving Biomolecule Activity
Controlling the degree of labeling is essential in Rhodamine conjugation. Excessive labeling may alter protein or antibody conformation and impair biological activity, while insufficient labeling can result in weak fluorescence signals. Optimizing reaction time, dye-to-biomolecule molar ratios, and buffer systems enables balanced labeling efficiency and signal intensity. Selecting appropriate conjugation strategies and mild reaction conditions helps preserve molecular function, ensuring compatibility with downstream applications such as live-cell imaging, protein interaction analysis, and high-throughput screening.
- Linker design: Introducing polyethylene glycol (PEG) linkers between Rhodamine and reactive groups increases aqueous solubility and provides spatial flexibility, reducing nonspecific interactions with protein surfaces and preventing self-quenching.
- Monitoring the F/P ratio (fluorophore-to-protein): UV–Vis spectroscopy is used to measure absorbance at 280 nm (protein) and ~550 nm (Rhodamine). Correction factors (CF) enable accurate calculation of the degree of labeling (DOL). Typically, a DOL of 2–4 provides optimal signal-to-noise performance.
- Purification strategies: After conjugation, products should be purified by gel filtration (desalting columns) or dialysis to remove unreacted free dye, ensuring that all detected fluorescence originates from true conjugates.
Targeted Research Applications Enabled by Rhodamine Labeling
Rhodamine fluorescence labeling provides powerful visualization and quantitative tools for biomolecular research, enabling detailed analysis of molecular behavior, protein dynamics, and cellular functions. In targeted research applications, Rhodamine labeling enhances signal sensitivity and is compatible with multiple imaging and analytical platforms, supporting complex experimental designs and precise data acquisition.
Fluorescence Imaging and Cellular Targeting Studies
Rhodamine-labeled molecules exhibit excellent performance in cellular imaging and targeting studies. Their high brightness and photostability allow prolonged observation of intracellular molecular behavior, while longer excitation and emission wavelengths reduce background autofluorescence. Using Rhodamine-labeled proteins, antibodies, or small-molecule probes, researchers can track intracellular distribution, dynamic transport, and subcellular localization, revealing signaling pathways, drug targeting efficiency, and organelle functions.
- Organelle-specific localization: Certain Rhodamine derivatives (e.g., Rhodamine 123) carry a positive charge and selectively accumulate in mitochondria driven by membrane potential, serving as live-cell indicators of apoptosis and metabolic state.
- High-resolution dynamic tracking: Rhodamine-labeled ligands enable visualization of receptor-mediated endocytosis in live cells. Exceptional photostability supports time-lapse imaging over several hours without significant photobleaching, which is critical for capturing vesicular transport dynamics.
Protein Localization and Trafficking Analysis
Protein localization and trafficking analysis is fundamental to understanding cellular biology. Rhodamine-labeled antibodies or fusion proteins enable precise subcellular localization without disrupting protein function. Combined with live-cell imaging, researchers can dynamically monitor protein endocytosis, exocytosis, nucleocytoplasmic transport, and membrane protein recycling. These studies are highly relevant to signaling pathway analysis, disease mechanism research, and development of therapeutic intervention strategies.
- Mechanistic studies of protein transport: Rhodamine labeling allows real-time quantitative analysis of protein translocation rates from cytoplasm to nucleus. Photo-activatable Rhodamine enables localized activation, allowing researchers to track intracellular diffusion dynamics.
- Super-resolution microscopy (SRM) applications: In STED (stimulated emission depletion) microscopy, Rhodamine dyes—particularly Si-Rhodamine and Texas Red—are among the most effective fluorophores. Their high stimulated emission cross-sections enable spatial resolution beyond the diffraction limit, reaching 20–50 nm, and allow detailed visualization of cytoskeletal microtubule structures.
Multiplex Fluorescence Assays and Co-localization Studies
Rhodamine dyes offer inherent advantages in multiplex fluorescence experiments. Their stable optical properties and relatively narrow emission spectra enable simultaneous use with other fluorophores for multichannel detection. By labeling different targets with Rhodamine, researchers can perform protein co-localization studies, complex analysis, and multi-target signal integration.
- Spectral complementarity: Rhodamine dyes (e.g., TRITC) pair seamlessly with FITC (green) and DAPI (blue). With emission peaks at 570–610 nm and minimal overlap with blue–green channels, Rhodamine provides high spatial accuracy in co-localization analysis.
- Multi-target co-detection: Combining Rhodamine-labeled antibodies with fluorescence in situ hybridization (FISH) probes allows simultaneous visualization of protein expression and genomic localization within the same tissue section.
Binding Kinetics and Interaction Mapping in Targeted Research
Rhodamine-labeled molecular probes enable precise measurement of binding kinetics and interaction network mapping. By monitoring fluorescence changes during association and dissociation, researchers can obtain accurate binding constants, affinities, and kinetic parameters. These applications are essential for receptor–ligand studies, enzyme–substrate recognition, and protein–protein interaction analysis. Coupled with high-sensitivity detection platforms, Rhodamine labeling supports interaction studies of low-abundance molecules, early signal detection, and high-throughput screening.
- FRET-based interaction detection: Rhodamine dyes (e.g., Rhodamine B) are commonly used as acceptors in fluorescence resonance energy transfer (FRET) paired with donors such as FITC. When the distance between two labeled molecules is less than 10 nm, energy transfer indicates direct molecular interaction.
- Fluorescence polarization (FP) and kinetics: Due to moderate excited-state lifetimes and high fluorescence intensity, Rhodamine-labeled small-molecule ligands are well suited for fluorescence polarization assays. Changes in polarization enable precise determination of binding affinities (K_d), widely used in high-throughput drug screening.
- Fluorescence correlation spectroscopy (FCS): Leveraging Rhodamine's single-molecule sensitivity at extremely low concentrations, FCS analyzes molecular diffusion coefficients in solution, allowing estimation of hydrodynamic radii and binding states with partner proteins.
Key Technical Considerations in Custom Rhodamine Labeling
When performing Rhodamine fluorescence labeling experiments, in addition to selecting appropriate conjugation chemistry, several critical technical factors must be considered. These factors directly influence labeling efficiency, fluorescence signal quality, and biomolecular activity, ultimately determining the reliability and reproducibility of experimental results. Scientifically designed experimental workflows and optimization strategies can maximize labeling performance and ensure effective application of Rhodamine labeling in complex biological systems.
Optimizing Labeling Efficiency for Low-Abundance Targets
Fluorescence labeling of low-abundance biomolecules represents a core challenge in high-sensitivity detection. Labeling efficiency can be optimized by adjusting the dye-to-target molar ratio, reaction time, pH, and buffer systems. For proteins and antibodies, stepwise dye addition or low-temperature reactions can improve labeling uniformity and reduce side reactions. For nucleic acids or small-molecule probes, selecting highly reactive Rhodamine derivatives can significantly enhance conjugation efficiency. Efficient labeling ensures that low-abundance targets still generate detectable signals, providing a reliable basis for quantitative analysis and kinetic studies.
- Signal amplification strategies: When direct labeling does not meet sensitivity requirements, biotin–streptavidin systems are commonly employed. By decorating streptavidin carriers with multiple Rhodamine molecules, signal amplification by orders of magnitude can be achieved.
- Reaction kinetics optimization: For extremely low substrate concentrations, precise adjustment of ionic strength, temperature, and controlled perturbations (e.g., microfluidic mixing) can increase collision frequency between reactive Rhodamine derivatives and targets, thereby improving grafting efficiency without increasing background signals.
Minimizing Non-Specific Binding and Background Fluorescence
The polycyclic aromatic structure of Rhodamine dyes confers inherent hydrophobicity, which can readily lead to nonspecific adsorption to membrane lipids or off-target proteins. Nonspecific binding and background fluorescence are major factors affecting signal-to-noise ratios. Common strategies include selecting highly specific conjugation chemistries, optimizing buffer systems to reduce nonspecific adsorption, purifying target molecules prior to labeling, and incorporating appropriate blocking agents during experiments. In cellular imaging applications, proper washing and blocking steps can significantly reduce background fluorescence. Reducing nonspecific signals enables higher-precision quantitative analysis and molecular localization studies.
- Charge modulation and chemical modification: Introducing sulfonate groups (sulfonation) into the Rhodamine scaffold can significantly enhance water solubility. Negatively charged sulfonated Rhodamine dyes (e.g., structures analogous to Alexa Fluor 594) effectively reduce nonspecific interactions with negatively charged cell membranes through electrostatic repulsion.
- Blocking strategies: In antibody-based detection, the use of highly optimized blocking buffers containing specific detergents and irrelevant proteins is critical for minimizing background fluorescence. In some cases, inclusion of trace amounts of organic solvents (e.g., 1% DMSO) during wash steps can help remove physically adsorbed free Rhodamine.
Photostability and Signal Consistency in Long-Term Experiments
Fluorescent dyes may undergo photobleaching during prolonged observation or repeated excitation. Although Rhodamine dyes are well known for their excellent photostability, careful experimental design is still required to maintain signal consistency. Common approaches include using antifade reagents, adjusting excitation intensity, employing short exposure times, and implementing interval-based image acquisition. For dynamic and high-resolution imaging, stable fluorescence signals are essential to ensure experimental reproducibility and data reliability.
- Photophysical stabilization: Photobleaching is often caused by reactions between excited-state fluorophores and molecular oxygen, generating singlet oxygen. Adding antioxidants (e.g., reduced glutathione) or commercial antifade reagents to the experimental system can significantly extend the fluorescence lifetime of Rhodamine dyes.
- Structural reinforcement strategies: Modern chemical modification techniques introduce rigid cyclic substituents (e.g., azacyclic structures) at the amino positions of Rhodamine, effectively restricting vibrational energy levels and markedly increasing total photon yield. This ensures consistent fluorescence signals under prolonged continuous laser excitation.
Compatibility with Downstream Analytical and Imaging Platforms
Rhodamine-labeled molecules must be compatible with a wide range of downstream analytical and imaging platforms, including confocal microscopy, flow cytometry, fluorescence plate readers, and multiplex fluorescence detection systems. When designing labeling experiments, factors such as excitation/emission spectra, fluorescence intensity, photobleaching rates, and potential fluorescence resonance energy transfer (FRET) effects must be considered. Through appropriate selection of labeling strategies and optimization of experimental conditions, Rhodamine-labeled biomolecules can achieve high-quality detection and reliable quantification across different platforms, supporting multidimensional research applications.
- Accurate calculation of spectral overlap: In multicolor experimental design, the red-tail emission of Rhodamine dyes must be considered. The use of narrow band-pass filters or spectral unmixing algorithms (linear unmixing) can effectively eliminate bleed-through between fluorescence channels.
- Compatibility with mass spectrometry and chromatography: In pharmacokinetic studies, labeled small-molecule probes often require validation by HPLC or LC–MS. It is essential to ensure that polarity changes introduced by Rhodamine labeling do not significantly alter chromatographic retention times and that the conjugation bond remains stable under electrospray ionization (ESI) conditions without fragmentation.
Custom Rhodamine Labeling Services at BOC Sciences
BOC Sciences provides comprehensive custom Rhodamine labeling services, delivering high-precision, high-stability, and reproducible labeling solutions for research involving proteins, antibodies, peptides, and nucleic acids. Leveraging advanced conjugation technologies and extensive expertise in fluorescence chemistry, we offer personalized labeling strategies tailored to specific research needs, ensuring reliable experimental outcomes and superior visualization performance.
High-Efficiency Rhodamine Labeling Services for Proteins
- Multiple conjugation strategies are available, including NHS ester and maleimide chemistries, enabling highly selective labeling of lysine and cysteine residues on proteins.
- Precise control of dye-to-protein molar ratios to preserve native protein conformation and biological activity.
- Post-labeling purification services, including dialysis, gel filtration, or column chromatography, to ensure impurity removal and labeling uniformity.
- Detailed product performance validation reports, including labeling efficiency, fluorescence intensity, and stability data, to facilitate rapid evaluation of experimental feasibility.
Custom Rhodamine-Labeled Antibody Services
- Support for Rhodamine conjugation of full-length antibodies and antibody fragments (Fab, scFv), suitable for immunofluorescence, flow cytometry, and imaging applications.
- Optimized buffer systems and conjugation conditions to maximize retention of antibody binding capacity and specificity.
- Post-labeling purification and functional validation, including SDS-PAGE and binding activity assays, to ensure reliable research data.
- Customization of fluorescence wavelengths and photostability to accommodate multicolor labeling and complex experimental designs.
Precision Rhodamine Labeling Services for Peptides
- Support for selective conjugation of a wide range of peptides at the N-terminus, C-terminus, or specific side-chain residues.
- Adjustable labeling ratios and reaction conditions to achieve optimal efficiency while maintaining peptide structure and functionality.
- Purification and validation of labeled peptides, including HPLC separation and mass spectrometric identification, ensuring high-purity products for downstream applications.
- Custom design of multiplex labeling or specialized fluorescence combinations to meet high-throughput screening or multiparametric detection requirements.
High-Precision Rhodamine Labeling Services for Nucleic Acids
- Support for selective 5′ or 3′ end labeling of single-stranded DNA, RNA, and oligonucleotides, preserving sequence integrity and stability.
- Optimized buffer systems and reaction conditions to ensure high and reproducible labeling efficiency.
- Purification and performance validation of labeled nucleic acids, including PAGE, HPLC, and fluorescence characterization, ensuring experimental reliability.
- Custom multicolor labeling solutions to support multiplex detection, real-time imaging, and molecular interaction analysis.
Do You Need A Consultation?
BOC Sciences integrates cutting-edge fluorescence technologies to accelerate your research, driving next-generation solutions for drug discovery and diagnostics.
Transform Your Studies with Cutting-Edge Fluorescent Products
| Cat. No. | Product Name | CAS No. | Inquiry |
|---|---|---|---|
| A14-0036 | Rhodamine B hydrazide | 74317-53-6 | Bulk Inquiry |
| F05-0031 | 6-Carboxy-X-rhodamine | 194785-18-7 | Bulk Inquiry |
| A17-0069 | Rhodamine 590 Chloride | 3068-39-1 | Bulk Inquiry |
| A17-0106 | Rhodamine 19 Perchlorate | 62669-66-3 | Bulk Inquiry |
| A16-0014 | Sulforhodamine 101 | 60311-02-6 | Bulk Inquiry |
| A17-0061 | Rhodamine 610 Perchlorate | 23857-51-4 | Bulk Inquiry |
| A03-0012 | Dihydrorhodamine 123 | 109244-58-8 | Bulk Inquiry |
| A17-0047 | Rhodamine 700 perchlorate | 63561-42-2 | Bulk Inquiry |
| A16-0142 | Dihydrorhodamine 6G | 217176-83-5 | Bulk Inquiry |
High-Performance Fluorescent Tools for Your Research
- BODIPY Bright, stable dyes used in bioimaging and molecular labeling.
- Coumarin Blue-emitting dyes for chemical sensing and fluorescence studies.
- Cyanine Versatile dyes used in biosensing and nucleic acid detection.
- Fluorescent Protein Used for live-cell imaging and real-time biosensing.
More About Rhodamine Dyes
Online Inquiry